Research
Background
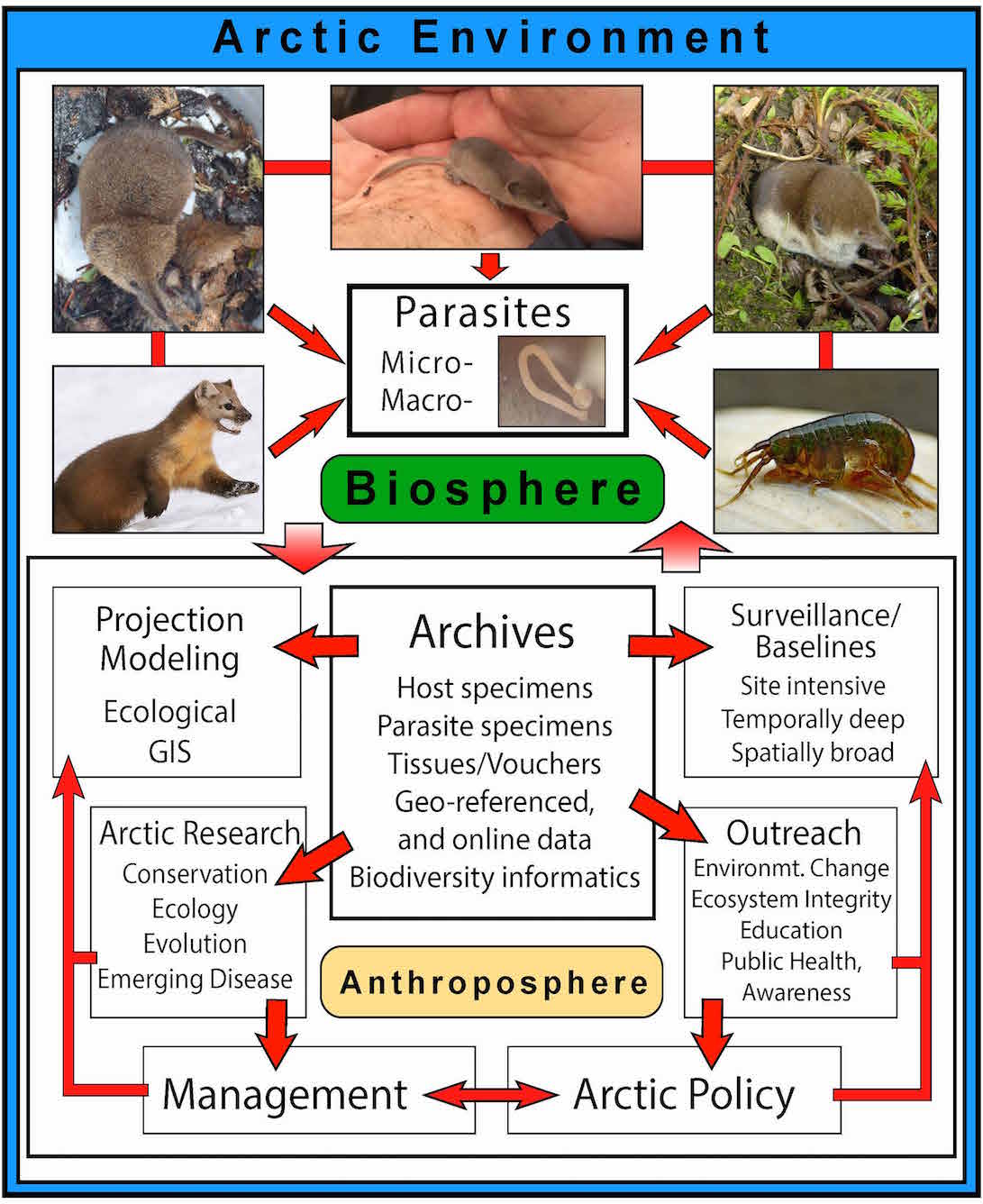 My background is in mammalian biogeography and molecular ecology. These disciplines constitute an essential foundation for understanding biodiversity responses to contemporary environmental change. My research strongly emphasizes integration among major disciplines (ecology and evolution) and among interrelated taxonomic groups (mammalian hosts and their parasites) for better understanding the complexity of diversification, demographic change, and interactions among disparate groups of organisms through time. In particular, I investigate mammalian host interactions across ecotones (transition zones where communities associated with distinct habitats interact). Within these boundary regions, animals may experience both gene flow and competition, influencing their evolutionary trajectories, fitness, relative susceptibility to infection by parasites, and regional variation in community assembly. I integrate a suite of traditional methods in mammalian biology (e.g., morphology and amplicon sequencing) with emerging genomic and stable isotope methods to test hypotheses of functional changes that impact community dynamics, ultimately with an aim to conserve intact wild systems. As such my research has a strong applied component towards biodiversity conservation, management under future global change scenarios, and emerging human/wildlife disease, fitting well within the Fisheries Wildlife and Conservation program at K-State.
My background is in mammalian biogeography and molecular ecology. These disciplines constitute an essential foundation for understanding biodiversity responses to contemporary environmental change. My research strongly emphasizes integration among major disciplines (ecology and evolution) and among interrelated taxonomic groups (mammalian hosts and their parasites) for better understanding the complexity of diversification, demographic change, and interactions among disparate groups of organisms through time. In particular, I investigate mammalian host interactions across ecotones (transition zones where communities associated with distinct habitats interact). Within these boundary regions, animals may experience both gene flow and competition, influencing their evolutionary trajectories, fitness, relative susceptibility to infection by parasites, and regional variation in community assembly. I integrate a suite of traditional methods in mammalian biology (e.g., morphology and amplicon sequencing) with emerging genomic and stable isotope methods to test hypotheses of functional changes that impact community dynamics, ultimately with an aim to conserve intact wild systems. As such my research has a strong applied component towards biodiversity conservation, management under future global change scenarios, and emerging human/wildlife disease, fitting well within the Fisheries Wildlife and Conservation program at K-State.
My research is highly specimen-based, relying on museum resources that have been built by past generations of wildlife biologists to vouch for the existence of a given species or set of species, at a given time and place. In addition to use of existing archives, I advocate for ethical expansion of these resources into the future, building knowledge and continuity for increasingly effective biodiversity conservation. My research therefore incorporates field expeditions to sample study species from sometimes extremely remote areas, filling in knowledge gaps of species distributions and reducing "data deficiency" as a factor that still influences biodiversity management, even for mammals. In addition to maximizing spatial sampling, I maintain a subset of sites for repeat sampling to build time-series of specimens that vouch for ecological and evolutionary change in response to local, regional, and global environmental shifts. As such, long-term research sites offer valuable opportunities for gathering robust physical biotic resources to increase the impact of temporal datasets.

If you haven't noticed yet, my primary research taxa are shrews! But, shrews don't live in a vacuum, and my publications reflect a broader focus on shrews within their respective small mammal communities, whole-community responses to changing climate variables across regional landscapes, and what can be learned by studying their diverse parasite faunas with highly variable host specificity. Shrews are a great group to study from an evolutionary perspective because they are diverse (at least for mammals), occupy most available terrestrial habitats, are highly fecund with short generation times, and along with tiny size and fast metabolism, these traits translate to relatively rapid evolutionary rates. This leads to fine scales of inference for processes of divergence, and demographic and functional changes. Shrews are also host to extremely diverse yet understudied parasite faunas. These diminutive mammals are "insectivores" feeding on arthropods and molluscs, which together constitute the primary and intermediate hosts necessary for completing complex helminth parasite lifecycles. Above all, shrews are just really cool...
Broad Research Areas
Phylogenetic Systematics, Phylogeography, and Conservation Genetics
These disciplines very generally focus on understanding evolutionary relationships across different spatial and temporal scales from global to local, and from deep to shallow time, but they are not mutually exclusive, and any study emphasizing one of these fields would benefit from more inclusive interpretation. Fundamentally, understanding the evolutionary relationships among families, genera, and species (systematics) will help us explain patterns of intra-specific diversification across a geographic range in response to environmental processes of change (phylogeography), and strengthen our ability to diagnose and effectively manage threatened populations (conservation genetics).
My earlier studies consisted of single or multi-species phylogeographic assessments to identify distinct evolutionary lineages within species and understand the geographic extent of these lineages, how they were influenced by climate and physical landscape features (e.g., isolating barriers) and when all of the major changes took place. For example, if a warming trend 130 thousand years ago (during the Sangamon interglacial) caused a particular lineage to expand its range northward in western North America, and this pattern was repeated for western lineages among multiple species, and again during the Holocene warming transition just 10 thousand years ago, then we might begin to infer what effect continued contemporary global warming might have on species that respond one way or another.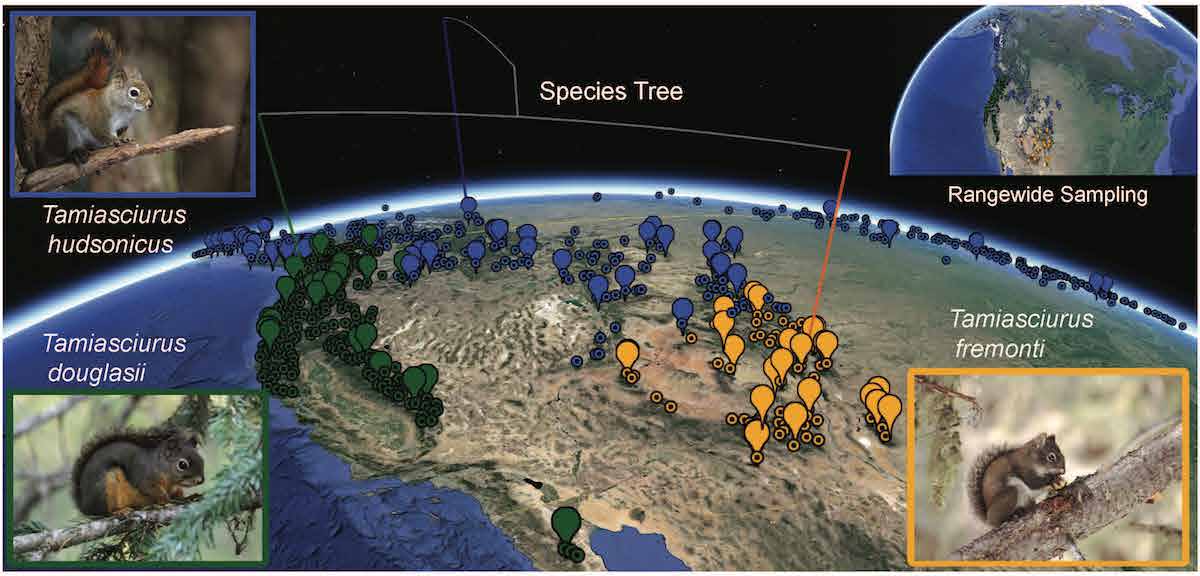
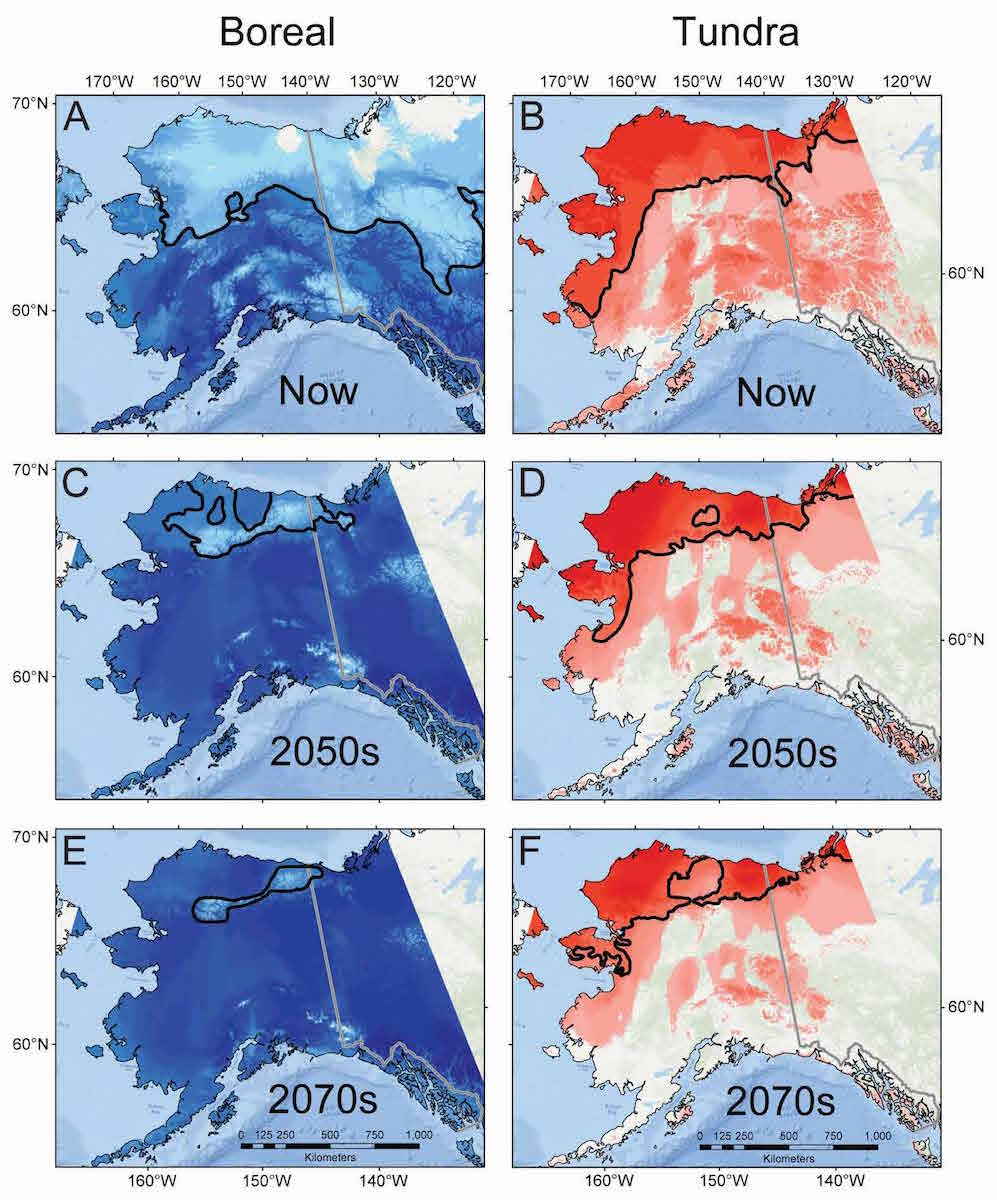
Each independent phylogeographic study adds pieces of the puzzle about how total biodiversity responds through time to change, and I have contributed several pieces, with more in progress! Congruence of pattern and process inferred across independent studies is now increasingly being solidified as analytical techniques advance by combining data from multiple species in comparative analyses. The major benefit beyond increased statistical rigor, is that we can now identify hotspots of genetic diversity, or areas at particular risk of losing distinct genetic diversity across entire communities (maps below: indicate projected trends in forest and tundra small mammal diversity into the near future). The reasons for increased risk are many, but include in particular human land practices (development, industry, agriculture), the combined attributes of climate change (both background and anthropogenic) and geography, and complex interactions between populations and species occupying a given area, such as gene flow through hybridization, competition, and transfer of parasites and pathogens.
 A good example of integration (systematics <--> phylogeography <--> conservation) from my own research considers the evolutionary history of the Sorex cinereus complex of shrews, consisting of >12 species of shrew distributed broadly across North America and Siberia. This group has diverged rapidly in response to late Quaternary climate cycling coupled with geographic complexity on a continental scale. Plylogenetic tree-based methods have been integral to understanding basic systematic relationships among the species, some of which are still unclear and under continued study. Phylogeographic analyses have highlighted the importance of climate and geography for diversification, and have identified multiple distinct lineages and isolated populations, some that have not been formally described, but that warrant conservation as distinct but threatened genetic pools.
A good example of integration (systematics <--> phylogeography <--> conservation) from my own research considers the evolutionary history of the Sorex cinereus complex of shrews, consisting of >12 species of shrew distributed broadly across North America and Siberia. This group has diverged rapidly in response to late Quaternary climate cycling coupled with geographic complexity on a continental scale. Plylogenetic tree-based methods have been integral to understanding basic systematic relationships among the species, some of which are still unclear and under continued study. Phylogeographic analyses have highlighted the importance of climate and geography for diversification, and have identified multiple distinct lineages and isolated populations, some that have not been formally described, but that warrant conservation as distinct but threatened genetic pools.
My colleagues and I have now amassed multiple lines of evidence for updating the taxonomy of the Sorex cinereus complex, investigating how gene flow has influenced the process of diversification (see below), identifying critical geographic regions for genetic diversity and conservation potential that can be extended across many groups of species, and beginning to understand how parasite communities both influence and respond to changes in these shrew hosts. Data include amplicon sequences from mtDNA and nuDNA (Hope et al. 2012), and soon to be published morphometric data, >100 mitogenomes, 20 microsatellite loci over >700 specimens representing all species, and a RadSeq dataset in progress (map below: indicates shrew relationships and geographic sampling coverage across species of the cinereus group). Finally, years of fieldwork to obtain shrew samples have yielded extensive collections of ecto- and endoparasites that are revealing new species, new locality records, and new host associations.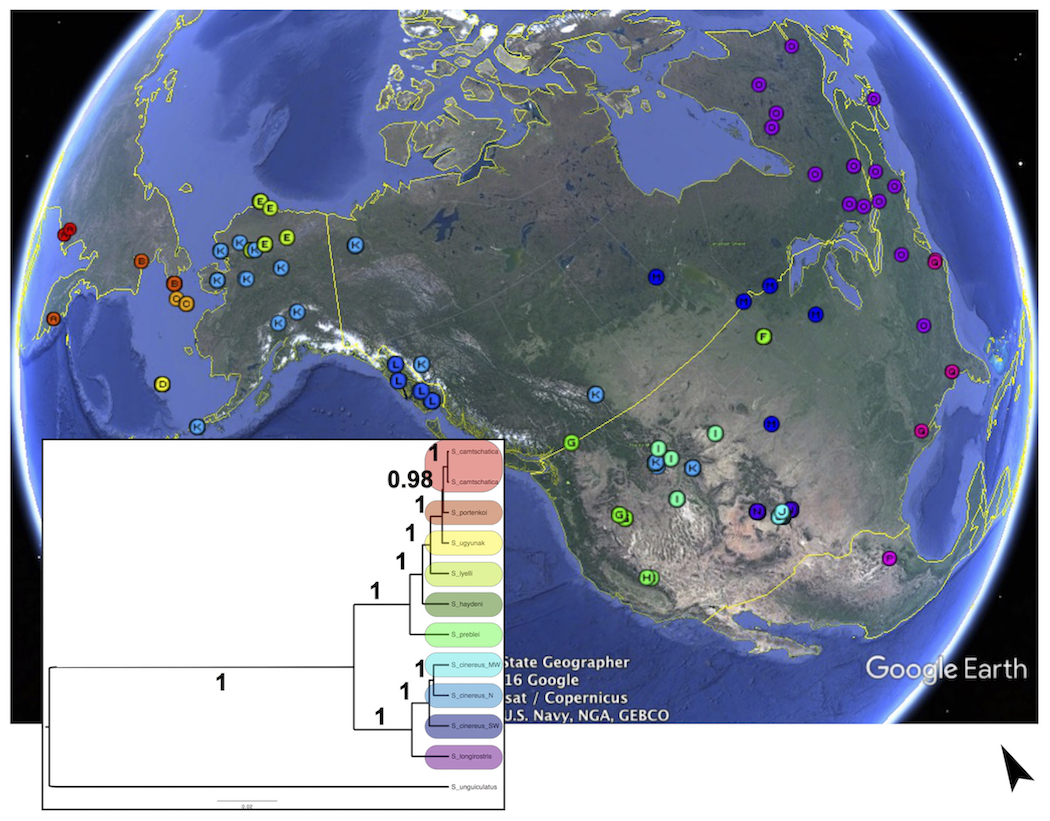
Hybridization, Speciation, and Host-parasite Co-evolution
One of the highest impacts of many phylogeographic studies (including my own) is through diagnosis of geographic zones that are congruent with common evolutionary discontinuities both within and among many species of mammals, birds, parasites etc. These "suture zones" often occur either across major geographic features, or across ecological transitions between distinct habitats or biomes. Where transitions are not absolute physical barriers, there are varying degrees of biotic mixing and interaction. Intra-specific hybridization is increasingly considered an evolutionary driver that may promote or suppress the formation of new species. Gene flow among more diverged species also occurs commonly, and introduction or rearrangement of genomic elements can have functional consequences for continued adaptive change, and for inter-dependent biodiversity, such as affecting the relative susceptibility of shrews to infection by parasites.
My research is increasingly focused on understanding the (co)evolutionary dynamics surrounding zones of contact between distinct communities. While working with the U.S. Geological Survey at the Alaska Scieince Center in Anchorage (2010-2014), I helped to develop microsatellite loci for shrews that for the first time have demonstrated hybridization between sibling species across the boreal forest-Arctic tundra ecotone in northern Alaska. The large microsat dataset that I have built in the last few years encompasses robust specimen series of both Sorex cinereus (sensu stricto) and S. ugyunak across this zone of contact for a first assessment of hybridization dynamics. Preliminary analyses are underway, and publications soon to follow... In addition to ongoing hybridization, it appears that gene flow has been a long term contributor to rapid speciation within this group.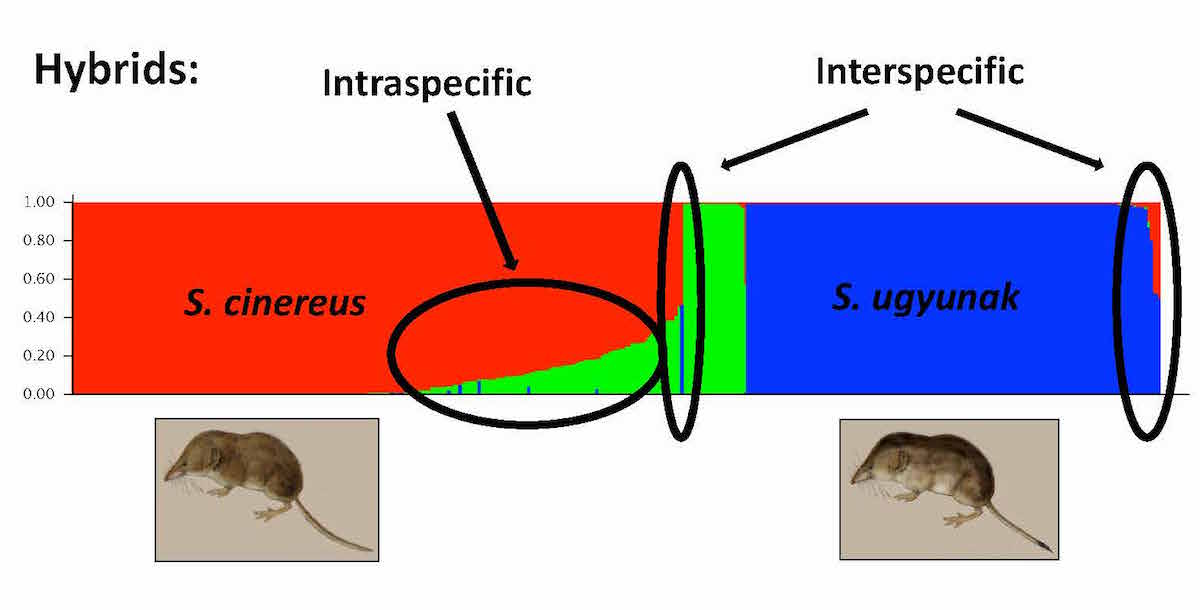
In the last 10 years, I have been working closely with U.S. government agencies administering public lands through northern Alaska to access remote areas across the boreal-tundra ecotone and perform extensive small mammal and parasite sampling. The Arctic is experiencing some of the most rapid rates of climate induced biotic changes globally, and as climates warm, entire communities of Arctic organisms are experiencing dynamic shifts in distributions, population demographics, and species interactions. Collections based on these ongoing field expeditions will be a vital resource for tracking rates and extent of change. Agencies collaborations include the National Park Service Arctic Network, U.S. Fish and Wildlife Arctic National Wildlife Refuge, Alaska Department of Fish and Game, and the U.S. Geological Survey Alaska Science Center.
More recently, at Kansas State University, I have shifted partial focus to the Mid-West U.S., implementing the same overarching research program across the mid-continental transition between Great Plains grasslands and eastern deciduous forests. This major ecotone stretches from northern Manitoba south through Texas. The Great Plains essentially constitute a filter zone for biodiversity between western and eastern North America, and yet they are also one of the most heavily human disturbed ecosystems worldwide, including development, agriculture, industry, and interruption of natural processes that historically maintained native prairies, including loss of mega-herbivores, and wildfire suppression. My prediction is that this fragmentation of previously continuous prairie has made the Great Plains biological filter more permeable to species movements between west and east, promoting increased interactions between distinct prairie and woodland communities. This system is parallel from a theoretical and functional standpoint to the boreal-tundra ecotone in the Arctic, and these combined regions now form the basis of my ongoing research program.
Small mammal trophic dynamics and community evolutionary ecology
Another benefit working at K-State is the proximity of the Konza Prairie Biological Station which has hosted the Konza Long Term Ecological Research (LTER) Program for over 40 years, and more recently a core National Ecological Observatory Network (NEON) site. I have adopted the small mammal long-term sampling transects here, partly to maintain long-term data continuity, and also to advance my own research. Konza will constitute the main long-term sampling site along the Great Plains-eastern forests ecotone (mentioned above) and is of high potential for understanding in greater detail the interactions among communities of small mammals and their parasites that are associated with prairie or woodland habitats. Konza is a landscape scale experiment of grazing and fire treatments and how these variables impact native prairie habitats.
While practicing both catch-and-release sampling and specimen removal, I am building robust specimen series from Konza that are being used for multiple studies. In a recent study (Hope et al. 2018) we showed that reasonable specimen sampling had no long-term impact on populations compared to non-removal sites. But, one major caveat was that these data were from a single ecosystem in the Southwest US. I hope to show that our findings are robust across different sites, such as Konza, and demonstrate the value of having specimen collections for long-term and integrated research as opposed to purely release trapping methods.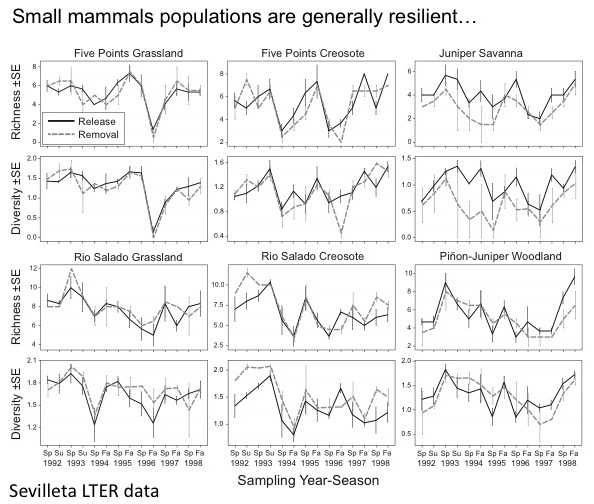
The major impact of these archives is not to assess the consequences of removing specimens, but to use the specimen parts for broader research application. Most recently, an undergraduate in my lab, Bia Gragg, performed a stable isotope analysis of two years of small mammal samples from Konza to investigate the role of small mammals as consumers on Konza and begin to understand community and food web dynamics among these vertebrates. Bia has demonstrated that isotopic signatures from fur (indicating spring diet, following molt) suggest significant overlap among all small mammals, whereas signatures from liver tissue (reflecting summer diet from the time of specimen collection) suggest dietary partitioning. Added complexity includes seasonal, inter-annual, and inter-specific variability, as well as intra-specific variability for populations of a given species occupying different habitat types. A cool story that was recently published in Ecosphere! For a summary, see Bia's ASM2018 poster here...
An added benefit of having full specimens, is that we gain all of the parasite materials, and confirmed associations between a particular mammal and a particular parasite. Agustin Jimenez at Southern Illinois University is currently identifying the helminth parasites from the first two years of data collection and has discovered two new nematode species, new locality records, and unique genetic diversity among these Konza worms. A first glimpse at the total diversity within the local small mammals. See Agustin's ASM2018 poster here... In addition, tick borne pathogens are on the rise in Kansas, and yet host and vector associations are still poorly understood. We have a growing collecting of preserved ticks pulled from now preserved mammal specimens. Both ticks and host tissues can be tested for pathogens, facilitating multiple future research avenues. As an example, I sent tissues samples of Blarina shrews collected from Konza to colleagues at Old Dominion University. Tests for Lyme's disease indicated these shrews were positive for Borrelia burgdorferi, a potential paradigm shift from an assumption that Peromyscus mice and Ixodes ticks are the major host and vector, respectively.
I state above "community evolutionary ecology" because it is increasingly evident that a deer mouse from western North America is not the same as one from the east in terms of their morphology, their evolutionary history, their habitat or disease ecology, etc... Yet, Kansas sits on shifting gradients of temperature and precipitation that have caused major distributional changes among local species over the past century. Ultimately, very limited field surveys through this time mean we don't have detailed knowledge of which genetic lineages of small mammals actually occur where, and this can have consequences for how they interact within their respective communities. In the past four years we have sampled small mammals and their parasites extensively across the Great Plains, including North Dakota in 2016, Manitoba in 2019, Kansas and southern Texas in 2020, and Kansas, South Dakota and Minnesota in 2021. Future state-wide field surveys accompanied by genetic identification and tree-based methods of inferring evolutionary relationships will be essential to more accurately understand the complexity of ecological community dynamics within this region. This also consitutes the MS thesis research of Mr. Tommy Herrera who is focusing on mammalian suture zones across the Great Plains.
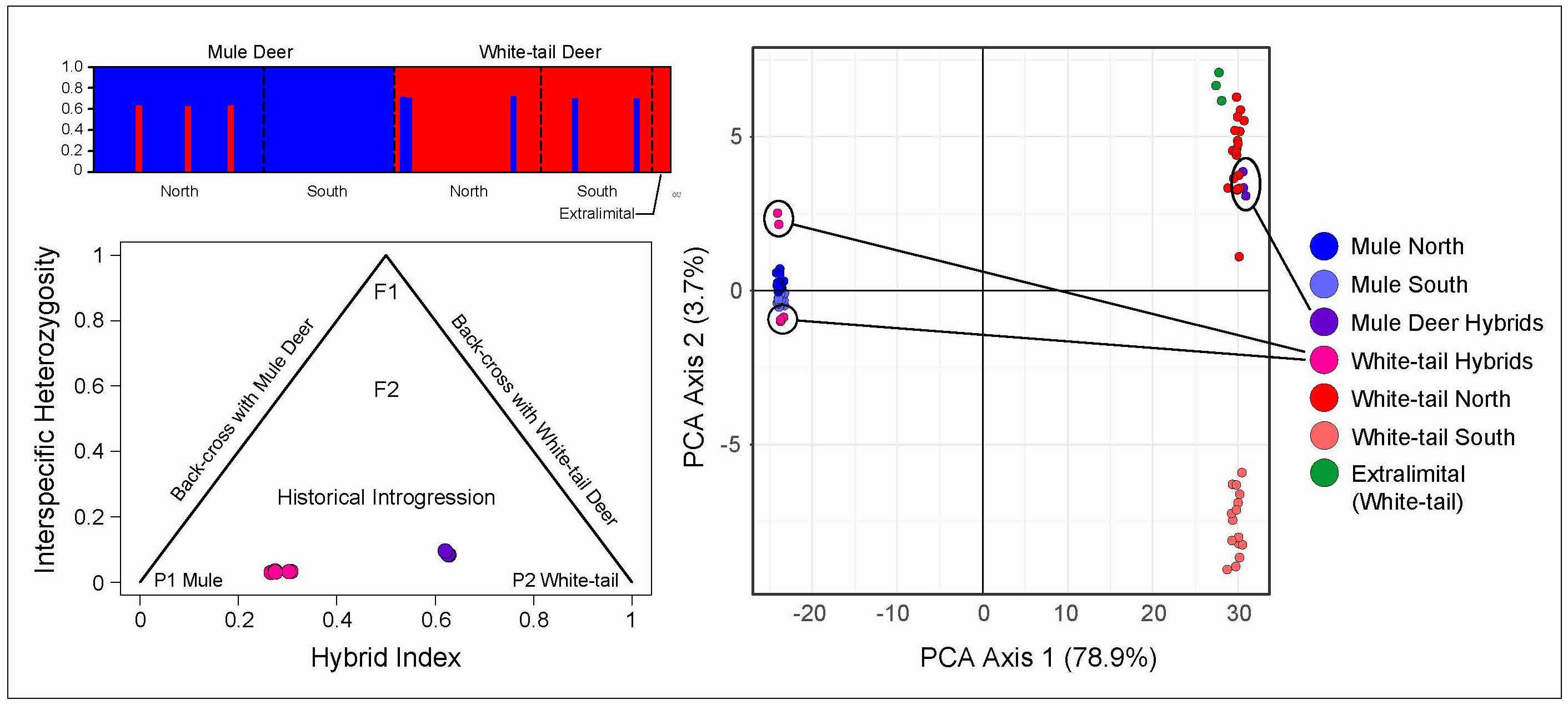
Finally, we were recently successful in being awarded funding from the USDA APHIS to continue our genomic studies of both mule and white-tailed deer across Kansas. This work has also been supported by the Kansas Department of wildlife and Parks. We have a preliminary publication recently released in Evolutionary Applications that shows roughly 10% of deer in western Kansas are of hybrid origin, with extensive and bi-directional backcrossing. A combination of ddRADseq data and mtDNA sequences strongly support a long history of peciation with gene flow among these deer, although conseqiences for the future integrity of these species, and for disease resistance is still less certain. We are now combining our assessments of landscape genomics and hybridization with genotyping of the PRNP (prion susceptibility) gene to diagnose the distribution of putative resistance to Chronic Wasting Disease across Kansas populations of deer.
Field Notes and Data Files
Manitoba July-August 2019 Field Notes
Expeditionary fieldwork across southern Manitoba to sample small mammals and parasites from multiple habitats across the Prairie to Eastern Forests ecotone. Field Notes
Chandalar River July-August 2017 Field Notes
Expeditionary fieldwork within the Arctic National Wildlife Refuge along the Junjik and Chandalar River corridors to sample small mammals and parasites across the Arctic Tundra to Boreal Forest ecotone. Field Notes