Source: Robert Szoszkiewicz, 785-532-0855, rs@phys.ksu.edu
Pronouncer: Szoszkiewicz is pronounced SHAWSH-kuh-vitz
Images available: Contact media@k-state.edu or call 785-532-6415
News release prepared by: Erinn Barcomb-Peterson, 785-532-6415, ebarcomb@k-state.edu
Thursday, July 2, 2009
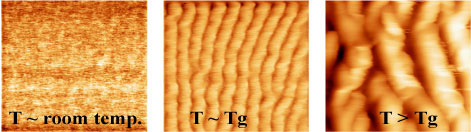 |
Examples of linear ripples obtained on a particular polymer as a result of scanning its surface with an atomic force tip heated to several different temperatures. Higher temperatures produced more pronounced ripples. (Image courtesy of K-State department of physics.) |
K-STATE PHYSICIST'S WORK WITH ATOMIC FORCE MICROSCOPE COULD LEAD TO TECHNOLOGIES FOR HEALTH CARE LIKE BETTER DIAGNOSTICS, DEEPER UNDERSTANDING OF CANCER
MANHATTAN -- Where biology, chemistry and physics intersect, a Kansas State University professor expects to find applications to improve human health.
Robert Szoszkiewicz, an assistant professor of physics at K-State, is continuing research on molecules both singularly and as a group. His study of proteins as a single molecule shows promise to help scientists understand the causes of diseases like some cancers. Meanwhile, his research on bunched molecules could lead to a more efficient way to identify antibodies in blood.
 |
Robert Szoszkiewicz |
Szoszkiewicz's research on proteins began at Columbia University and some of it was published in June in Nature Chemistry magazine. He and his former colleagues looked at the unexpected complexity in the dynamics of stretching and breaking of a single chemical bond between two atoms embedded in a protein. A cleavage of that particular bond has been precisely signaled by unfolding and elongation of a part of a protein.
"There is lots of potential for this research to really address issues of major importance in biology because this will be related to particular pathways through which some kinds of diseases, cancers and biological processes develop," he said. "And, we can study that on the very molecular level by studying the single proteins involved and stretching them and seeing how this contributes to the overall picture."
The research uses an atomic force microscope, a tool involving a cantilever with a sharp tip that under certain circumstances becomes attached to the protein molecule. The researchers stretch the molecules and measure precisely their tiny displacements.
"Any work that we do on the ground level of stretching any kind of chemical bonds is fundamentally important because it's the kind of knowledge no one has ever measured on this scale," he said. "Proteins constantly fold and unfold. A folded protein is the one that's biologically active and performing a function. So any problems during its folding and unfolding translate to some potential diseases like cancers."
Szoszkiewicz received a grant from K-State's Terry C. Johnson Center for Basic Cancer Research to use these techniques to stretch some proteins that are significant in breast cancer. He is collaborating with Anna Zolkiewska and Michal Zolkiewski, both K-State associate professors of biochemistry.
Another area of Szoszkiewicz's research began while at the Georgia Institute of Technology. It involves changing the properties of a surface at the very local scale by using an atomic force microscope in which the sharp cantilever tip can be heated up to between 500 degrees and 600 degrees Celsius.
When this heated tip is scraped across a surface, Szoszkiewicz said, the heat can -- under certain circumstances -- initiate a chemical reaction on that very surface. He and his colleagues have created patches of chemically changed surface only 10 to 15 nanometers across, but Szoszkiewicz thinks he will be able to change the surface just bunch of molecules at a time. This ability to pattern the surface, he said, could improve personalized medicine by allowing scientists to create tiny chips on which many tests can be run simultaneously.
"You could take a small sample of blood and screen it for any possible drug that could work for you," he said. "Ordinarily to do such a screening would require a humongous amount of testing material and antibodies. If you could prepare a surface that will selectively bind only one or two molecules of a kind and then see if you have bound or not, then you would need only a few milliliters of blood, and you could test it against millions of molecules. Of course, there are other factors involved and the particular interactions between single blood molecules and their antibodies might not be that simple. But this still remains to be seen."
Szoszkiewicz said that if scientists can pattern the surface in the way they like, another application might be tiny electric circuits and lenses that guide, respectively, electrons and photons. Also, using an atomic force microscope in this way, he said, physicists can not only change the chemical properties of the surface but they also can change the topography of a surface -- in other words, how the surface looks.
For example, Szoszkiewicz and colleagues have created and investigated the nature of nanoscale ripples created on polymers, or the "polymer dunes" at a nanoscale level. Using custom-made polymers, the researchers scratched them with the hot tip while using enough heat to indent within the polymer during scratching but not to destroy the polymer sample. Such research investigated how these tiny dunes created on polymers change with the application of heat. In this case, heat functions like wind would on real sand dunes. The researchers investigated under which conditions the dune gets pushed in the way they wanted it and forms a desired shape.
"This way you can prepare structures that can serve to sort materials at the nanoscale," he said. "Later on you virtually throw some other molecules on the surface, and maybe some of it will go into the grooves, depending on the chemistry."
The researchers were able to prepare several types of nanoscopic ripples -- these so-called tiny dunes -- with the most common type being the one in which the ripples organize in a roughly linear fashion. However, they also explored circular geometry. Their work has been published in the June issue of the journal Physical Review B.
"What we have proven in this paper is that, under certain conditions, by scanning continuously along these circles, you can make the ripples along your circular scanning path, and under certain circumstances to move them collectively and continuously," Szoszkiewicz said. "Measuring the collective velocities, we found that they would approach the velocities of shifting a big sand dune, just scaled down."
Szoszkiewicz's continuation of these projects in his lab at K-State include several other researchers: Heidi Martin, sophomore in physics from Junction City; Ashim Dey, doctoral student in physics, Manhattan; Neelam Khan, May 2009 doctoral graduate in physics; and Vera Okuneva, research assistant in biology.